Ploidy

 Clash Royale CLAN TAG#URR8PPP
Clash Royale CLAN TAG#URR8PPP 
Diploid cells have twice the number of chromosomes as haploid cells of the same species.
Ploidy (/ˈplɔɪdi/) is the number of complete sets of chromosomes in a cell, and hence the number of possible alleles for autosomal and pseudoautosomal genes.
Somatic cells, tissues and individuals can be described according to the number of sets present (the ploidy level): monoploid (1 set), diploid (2 sets), triploid (3 sets), tetraploid (4 sets), pentaploid (5 sets), hexaploid (6 sets), heptaploid[1] or septaploid[2] (7 sets), etc. The generic term polyploid is used to describe cells with three or more chromosome sets.[3][4]
Humans are diploid organisms, carrying two complete sets of chromosomes: one set of 23 chromosomes from their father and one set of 23 chromosomes from their mother. The two sets combined provide a full complement of 46 chromosomes. This total number of chromosomes is called the chromosome number. The zygotic number is defined as the number of chromosomes in zygotic cells. Human zygotes are diploid, hence with a zygotic number of 46.
When a species has a varying chromosome number, e.g. a diploid and tetraploid form, the chromosome number is called diploid number in the diploid form, and tetraploid number in the tetraploid form.
The number of chromosomes found in a single complete set of chromosomes is called the monoploid number (x). In most animals, the haploid number (n) is unique to gametes (sperm or egg cells), and refers to the total number of chromosomes found in a gamete, which under normal conditions is half the total number of chromosomes in a somatic cell.
The haploid number for humans (half of 46) is 23; and the monoploid number equals 46 divided by the ploidy level of 2, which is also 23. When a human germ cell undergoes meiosis the two sets of 23 chromosomes are split in half to form gametes. After fusion of a male and a female gamete (fertilization) both containing 1 set of 23 chromosomes, the resulting zygote has 46 chromosomes: 2 sets of 23 chromosomes (22 autosomes, and 1 allosome).
The common potato (Solanum tuberosum) is an example of a tetraploid organism, carrying four sets of chromosomes. The potato plant inherits two sets of 12 chromosomes from the pollen parent, and two sets of 12 chromosomes from the ovule parent. The four sets combined provide a full complement of 48 chromosomes. The haploid number (half of 48) is 24. The monoploid number equals the chromosome number divided by the ploidy level: 48 chromosomes in total divided by a ploidy level of 4 equals a monoploid number of 12.
The commercial common potato crop is propagated vegetatively (asexual reproduction through mitosis),[5] in which case new individuals are produced from a single parent, without the involvement of gametes and fertilization, and all the offspring are genetically equal to each other and to the parent.
| Ploidy | Nr of chromosomes | Somatic | Gametic |
|---|---|---|---|
| Diploid | 2x=22 | 2n=22 | n=11 |
| Tetraploid | 4x=44 | 2n=44 | n=22 |
| Hexaploid | 6x=66 | 2n=66 | n=33 |
| Octoploid | 8x=88 | 2n=88 | n=44 |
Because the chromosome number is generally reduced only by the specialized process of meiosis, the somatic cells of the body inherit and maintain the chromosome number of the zygote. However, in many situations somatic cells double their copy number by means of endoreduplication as an aspect of cellular differentiation. For example, the hearts of two-year-old children contain 85% diploid and 15% tetraploid nuclei, but by 12 years of age the proportions become approximately equal, and adults examined contained 27% diploid, 71% tetraploid and 2% octaploid nuclei.[6]
When a germ cell with an uneven number of chromosomes undergoes meiosis, the chromosomes can't be evenly divided between two cells resulting in aneuploid gametes. Triploid organisms for instance are usually sterile. Because of this, triploidy is a common way of making seedless fruit such as bananas and watermelons. If the fertilization of human gametes results in 3 sets of chromosomes the condition is called triploid syndrome.
Contents
1 Etymology
2 Special cases
3 Types of ploidy
3.1 Haploid and monoploid
3.2 Diploid
3.3 Homoploid
3.4 Zygoidy and azygoidy
3.5 Polyploidy
3.5.1 Polyploidy in bacteria and archaea
3.6 Variable or indefinite ploidy
3.7 Mixoploidy
3.8 Dihaploidy and polyhaploidy
3.9 Euploidy
4 Adaptive and ecological significance of variation in ploidy
5 Notes
6 References
7 Sources
8 External links
Etymology
The term ploidy is a back-formation from haploidy and diploidy. Ploid is a combination of Ancient Greek -παλτος (-paltos), -πλος (-plos), -πλόος (-plóos, "fold"), and -oid from Ancient Greek -ειδής (-eidḗs), -οειδής (-oeidḗs), from εἶδος (eîdos, "form, likeness").[a] The principal meaning of the Greek word ἁπλόος haplóos is "two-fold",[7] from ἅμα, which means, "at once, at the same time".[8] From this comes the secondary sense of "single", since folding double produces a unity. It is in this latter sense that it appears in modern genetics. διπλόος diplóos means "duplex" or "two-fold". Diploid therefore means "duplex-shaped" (compare 'humanoid', "human-shaped").
Eduard Strasburger, coined the terms haploid and diploid in 1905:
.mw-parser-output .templatequoteoverflow:hidden;margin:1em 0;padding:0 40px.mw-parser-output .templatequote .templatequoteciteline-height:1.5em;text-align:left;padding-left:1.6em;margin-top:0
Schließlich wäre es vielleicht erwünscht, wenn den Bezeichnungen Gametophyt und Sporophyt, die sich allein nur auf Pflanzen mit einfacher und mit doppelter Chromosomenzahl anwenden lassen, solche zur Seite gestellt würden, welche auch für das Tierreich passen. Ich erlaube mir zu diesem Zwecke die Worte Haploid und Diploid, bezw. haploidische und diploidische Generation vorzuschlagen.[9][10]
Some authors suggest that Strasburger based the terms on Weismann's conception of the id (or germ plasm),[11][12][13] hence haplo-id and diplo-id. The two terms were brought into the English language from German through William Henry Lang's 1908 translation of a 1906 textbook by Strasburger and colleagues.[14][citation needed]
Technically, ploidy refers to the nucleus. Though at times authors may report the total ploidy of all nuclei present within the cell membrane of a syncytium,[15] usually the ploidy of the nuclei present will be described. For example, a fungal dikaryon with two haploid nuclei is distinguished from the diploid in which the chromosomes share a nucleus and can be shuffled together.[16] Nonetheless, because in most situations there is only one nucleus, it is commonplace to speak of the ploidy of a cell.
Special cases
It is possible on rare occasions for ploidy to increase in the germline, which can result in polyploid offspring and ultimately polyploid species. This is an important evolutionary mechanism in both plants and animals.[17] As a result, it becomes desirable to distinguish between the ploidy of a species or variety as it presently breeds and that of an ancestor. The number of chromosomes in the ancestral (non-homologous) set is called the monoploid number (x), and is distinct from the haploid number (n) in the organism as it now reproduces. Both numbers n, and x, apply to every cell of a given organism.[citation needed]
Common wheat is an organism where x and n differ. It has six sets of chromosomes, two sets from each of three different diploid species that are its distant ancestors. The somatic cells are hexaploid, with six sets of chromosomes, 2n = 6x = 42 (where the monoploid number x = 7 and the haploid number n = 21). The gametes are haploid for their own species, but triploid, with three sets of chromosomes, by comparison to a probable evolutionary ancestor, einkorn wheat.[citation needed]
Tetraploidy (four sets of chromosomes, 2n = 4x) is common in plants, and also occurs in amphibians, reptiles, and insects. For example, species of Xenopus (African toads) form a ploidy series, featuring diploid (X.tropicalis, 2n=20), tetraploid (for example X.laevis, 4n=36), octaploid (for example X.wittei, 8n=72) and dodecaploid (for example X.ruwenzoriensis, 12n=108) species.[18]
Over evolutionary time scales in which chromosomal polymorphisms accumulate, these changes become less apparent by karyotype - for example, humans are generally regarded as diploid, but the 2R hypothesis has confirmed two rounds of whole genome duplication in early vertebrate ancestors.
Ploidy can also differ with life cycle.[19][20] In some insects it differs by caste. In humans, only the gametes are haploid, but in the Australian bulldog ant, Myrmecia pilosula, a haplodiploid species, haploid individuals of this species have a single chromosome, and diploid individuals have two chromosomes.[21] In Entamoeba, the ploidy level varies from 4n to 40n in a single population.[22]Alternation of generations occurs in many plants.
Some studies suggest that selection is more likely to favor diploidy in host species and haploidy in parasite species.[23]
Types of ploidy
Haploid and monoploid
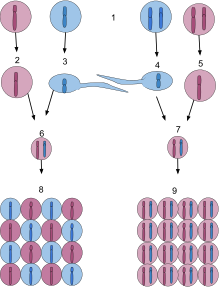
1.Haploid organism are on the left and Diploid organism on the right. 2.This is an haploid egg carrying the dominant purple gene. 3.This is a haploid sperm carrying the recessive blue gene. 4.This is a diploid sperm carrying the recessive blue gene. 5.This is an diploid egg carrying the dominant purple gene. 6.This is the short lived diploid state of haploid organisms. 7.This is the first stage of a zygote which has just been fertilized by a sperm. 8.The spores released by the diploid structure either express the mothers dominate gene or the fathers recessive gene. 9. The baby's cells express the dominant or recessive.
The nucleus of a eukaryotic cell is haploid if it has a single set of chromosomes, each one not being part of a pair. By extension a cell may be called haploid if its nucleus is haploid, and an organism may be called haploid if its body cells (somatic cells) are haploid. The number of chromosomes in a single set is called the haploid number, given the symbol n. If the number of chromosomes in the set is 1 (n=1) then the nucleus (or cell, organism) may be called monoploid.
Gametes (sperm and ova) are haploid cells. The haploid gametes produced by most organisms combine to form a zygote with n pairs of chromosomes, i.e. 2n chromosomes in total. The chromosomes in each pair, one of which comes from the sperm and one from the egg, are said to be homologous. Cells and organisms with pairs of homologous chromosomes are called diploid. For example, most animals are diploid and produce haploid gametes. During meiosis, sex cell precursors have their number of chromosomes halved by randomly "choosing" one member of each pair of chromosomes, resulting in haploid gametes. Because homologous chromosomes usually differ genetically, gametes usually differ genetically from one another.[citation needed]
All plants and many fungi and algae switch between a haploid and a diploid state, with one of the stages emphasized over the other. This is called alternation of generations. Most fungi and algae are haploid during the principal stage of their lifecycle, as are plants like mosses. Most animals are diploid, but male bees, wasps, and ants are haploid organisms because they develop from unfertilized, haploid eggs, while females (workers and queens) are diploid, making their system haplodiploid.
In some cases there is evidence that the n chromosomes in a haploid set have resulted from duplications of an originally smaller set of chromosomes. This "base" number – the number of apparently originally unique chromosomes in a haploid set – is called the monoploid number,[24] also known as basic or cardinal number,[25] or fundamental number.[26][27] As an example, the chromosomes of common wheat are believed to be derived from three different ancestral species, each of which had 7 chromosomes in its haploid gametes. The monoploid number is thus 7 and the haploid number is 3 × 7 = 21. In general n is a multiple of x. The somatic cells in a wheat plant have six sets of 7 chromosomes: three sets from the egg and three sets from the sperm which fused to form the plant, giving a total of 42 chromosomes. As a formula, for wheat 2n = 6x = 42, so that the haploid number n is 21 and the monoploid number x is 7. The gametes of common wheat are considered to be haploid, since they contain half the genetic information of somatic cells, but they are not monoploid, as they still contain three complete sets of chromosomes (n = 3x).[28]
In the case of wheat, the origin of its haploid number of 21 chromosomes from three sets of 7 chromosomes can be demonstrated. In many other organisms, although the number of chromosomes may have originated in this way, this is no longer clear, and the monoploid number is regarded as the same as the haploid number. Thus in humans, x = n = 23.
Diploid
Diploid cells have two homologous copies of each chromosome, usually one from the mother and one from the father. All or nearly all mammals are diploid organisms. The suspected tetraploid (possessing four chromosome sets) plains viscacha rat (Tympanoctomys barrerae) and golden vizcacha rat (Pipanacoctomys aureus)[29] have been regarded as the only known exceptions (as of 2004).[30] However, some genetic studies have rejected any polyploidism in mammals as unlikely, and suggest that amplification and dispersion of repetitive sequences best explain the large genome size of these two rodents.[31] All normal diploid individuals have some small fraction of cells that display polyploidy. Human diploid cells have 46 chromosomes (the somatic number, 2n) and human haploid gametes (egg and sperm) have 23 chromosomes (n).
Retroviruses that contain two copies of their RNA genome in each viral particle are also said to be diploid. Examples include human foamy virus, human T-lymphotropic virus, and HIV.[32]
Homoploid
"Homoploid" means "at the same ploidy level", i.e. having the same number of homologous chromosomes. For example, homoploid hybridization is hybridization where the offspring have the same ploidy level as the two parental species. This contrasts with a common situation in plants where chromosome doubling accompanies, or happens soon after hybridization. Similarly, homoploid speciation contrasts with polyploid speciation.[citation needed]
Zygoidy and azygoidy
Zygoidy is the state where the chromosomes are paired and can undergo meiosis. The zygoid state of a species may be diploid or polyploid.[33][34] In the azygoid state the chromosomes are unpaired. It may be the natural state of some asexual species or may occur after meiosis. In diploid organisms the azygoid state is monoploid. (see below for dihaploidy)
Polyploidy
Polyploidy is the state where all cells have multiple sets of chromosomes beyond the basic set, usually 3 or more. Specific terms are triploid (3 sets), tetraploid (4 sets), pentaploid (5 sets), hexaploid (6 sets), heptaploid[1] or septaploid[2] (7 sets) octoploid (8 sets), nonaploid (9 sets), decaploid (10 sets), undecaploid (11 sets), dodecaploid (12 sets), tridecaploid (13 sets), tetradecaploid (14 sets) etc.[35][36][37][38] Some higher ploidies include hexadecaploid (16 sets), dotriacontaploid (32 sets), and tetrahexacontaploid (64 sets),[39] though Greek terminology may be set aside for readability in cases of higher ploidy (such as "16-ploid").[37]Polytene chromosomes of plants and fruit flies can be 1024-ploid.[40][41] Ploidy of systems such as the salivary gland, elaiosome, endosperm, and trophoblast can exceed this, up to 1048576-ploid in the silk glands of the commercial silkworm Bombyx mori.[15]
The chromosome sets may be from the same species or from closely related species. In the latter case, these are known as allopolyploids (or amphidiploids, which are allopolyploids that behave as if they were normal diploids). Allopolyploids are formed from the hybridization of two separate species. In plants, this probably most often occurs from the pairing of meiotically unreduced gametes, and not by diploid–diploid hybridization followed by chromosome doubling.[42] The so-called Brassica triangle is an example of allopolyploidy, where three different parent species have hybridized in all possible pair combinations to produce three new species.
Polyploidy occurs commonly in plants, but rarely in animals. Even in diploid organisms, many somatic cells are polyploid due to a process called endoreduplication where duplication of the genome occurs without mitosis (cell division).
The extreme in polyploidy occurs in the fern genus Ophioglossum, the adder's-tongues, in which polyploidy results in chromosome counts in the hundreds, or, in at least one case, well over one thousand.
It is possible for polyploid organisms to revert to lower ploidy by haploidisation.
Polyploidy in bacteria and archaea
Polyploidy is a characteristic of the bacterium Deinococcus radiodurans [43] and of the archaeon Halobacterium salinarum.[44] These two species are highly resistant to ionizing radiation and desiccation, conditions that induce DNA double-strand breaks.[45][46] This resistance appears to be due to efficient homologous recombinational repair.
Variable or indefinite ploidy
Depending on growth conditions, prokaryotes such as bacteria may have a chromosome copy number of 1 to 4, and that number is commonly fractional, counting portions of the chromosome partly replicated at a given time. This is because under exponential growth conditions the cells are able to replicate their DNA faster than they can divide.
In ciliates, the macronucleus is called ampliploid, because only part of the genome is amplified.[47]
Mixoploidy
Mixoploidy is the case where two cell lines, one diploid and one polyploid, coexist within the same organism. Though polyploidy in humans is not viable, mixoploidy has been found in live adults and children.[48] There are two types: diploid-triploid mixoploidy, in which some cells have 46 chromosomes and some have 69,[49] and diploid-tetraploid mixoploidy, in which some cells have 46 and some have 92 chromosomes. It is a major topic of cytology.
Dihaploidy and polyhaploidy
Dihaploid and polyhaploid cells are formed by haploidisation of polyploids, i.e., by halving the chromosome constitution.
Dihaploids (which are diploid) are important for selective breeding of tetraploid crop plants (notably potatoes), because selection is faster with diploids than with tetraploids. Tetraploids can be reconstituted from the diploids, for example by somatic fusion.
The term "dihaploid" was coined by Bender[50] to combine in one word the number of genome copies (diploid) and their origin (haploid). The term is well established in this original sense,[51][52] but it has also been used for doubled monoploids or doubled haploids, which are homozygous and used for genetic research.[53]
Euploidy
Euploidy (Greek eu, true or even) is the state of a cell or organism having one or more than one set of the same set of chromosomes, possibly excluding the sex-determining chromosomes. For example, most human cells have 2 of each of the 23 homologous monoploid chromosomes, for a total of 46 chromosomes. A human cell with an extra set out of the 23 normal ones would be considered euploid. Euploid karyotypes would consequentially be a multiple of the haploid number, which in humans is 23. Aneuploidy is the state where one or more chromosomes of a normal set are missing or present in more than their usual number of copies. Unlike euploidy, aneuploid karyotypes will not be a multiple of the haploid number. In humans, examples of aneuploidy include having a single extra chromosome (such as Down syndrome), or missing a chromosome (such as Turner syndrome). Aneuploid karyotypes are given names with the suffix -somy (rather than -ploidy, used for euploid karyotypes), such as trisomy and monosomy.
Adaptive and ecological significance of variation in ploidy
A study comparing the karyotypes of endangered or invasive plants with those of their relatives found that being polyploid as opposed to diploid is associated with a 14% lower risk of being endangered, and a 20% greater chance of being invasive.[54] Polyploidy may be associated with increased vigor and adaptability.[55]
Notes
^ Compare the etymology of tuple, from the Latin for -fold.
References
^ ab U. R. Murty (1973). "Morphology of pachytene chromosomes and its bearing on the nature of polyploidy in the cytological races of Apluda mutica L". Genetica. 44 (2): 234–243. doi:10.1007/bf00119108.
^ ab Tuguo Tateoka (May 1975). "A contribution to the taxonomy of the Agrostis mertensii-flaccida complex (Poaceae) in Japan". Journal of Plant Research. 88 (2): 65–87. doi:10.1007/bf02491243.
^
Rieger, R.; Michaelis, A.; Green, M.M. (1976). Glossary of Genetics and Cytogenetics: Classical and Molecular (4th ed.). Berlin/Heidelberg: Springer-Verlag. p. 434. doi:10.1007/978-3-642-96327-8. ISBN 978-3-540-07668-1.
^ Darlington, C. D. (Cyril Dean) (1937). Recent advances in cytology. Philadelphia: P. Blakiston's son & co. p. 60.
^ "The Biology of Solanum tuberosum (L.) (Potatoes)". Canadian Food Inspection Agency.
^ John O. Oberpriller; A Mauro. The Development and Regenerative Potential of Cardiac Muscle. Taylor&Francis.
^ "Greek Word Study Tool". www.perseus.tufts.edu.
^ "Greek Word Study Tool". www.perseus.tufts.edu.
^ Strasburger, Eduard; Allen, Charles E.; Miyake, Kilchi; Overten, James B. (1905). "Histologische Beiträge zur Vererbungsfrage". Jahrbücher für wissenschaftliche botanik. Leipzig: Verlag von Gebrüder Borntraeger. 42: 62. Retrieved 2017-03-11.
^ Toepfer, Georg (2011). Historisches Worterbüch der Biologie - Geschichte und Theorie der biologischen Grundbegriffe. Stuttgart: J.B. Metzler'sche Verlagsbuchhandlung und Carl Ernst Poeschel Verlag GmbH. p. 169. ISBN 978-3-476-02317-9.
^ Battaglia E (2009). "Caryoneme alternative to chromosome and a new caryological nomenclature" (PDF). Caryologia. 62 (4): 48.
^ Haig David (2008). "Homologous versus antithetic alternation of generations and the origin of sporophytes" (PDF). The Botanical Review. 74 (3): 395–418. doi:10.1007/s12229-008-9012-x.
^ Bennett, Michael D. (2004). "Biological relevance of polyploidy: ecology to genomics". Biological Journal of the Linnean Society. London: The Linnean Society of London. 82 (4): 413. doi:10.1111/j.1095-8312.2004.00328.x.
^ Strasburger, E.; Noll, F.; Schenck, H.; Karsten, G. 1908. A Textbook of botany, 3rd English ed. (1908) [1], rev. with the 8th German ed. (1906) [2], translation by W. H. Lang of Lehrbuch der Botanik für Hochschulen. Macmillan, London.
^ ab Encyclopedia of the Life Sciences (2002) "Polyploidy" Francesco D'Amato and Mauro Durante
^ James B. Anderson; Linda M Kohn. "Dikaryons, diploids, and evolution" (PDF). University of Toronto.
^ Mable B. K. (2004). ""'Why polyploidy is rarer in animals than in plants': myths and mechanisms" in Biological relevance of polyploidy: ecology to genomics" (PDF). Biological Journal of the Linnean Society. 82: 453–466. doi:10.1111/j.1095-8312.2004.00332.x.
^ Schmid, M; Evans, BJ; Bogart, JP (2015). "Polyploidy in Amphibia". Cytogenet. Genome Res. 145 (3–4): 315–30. doi:10.1159/000431388. PMID 26112701.
^ Parfrey LW, Lahr DJG, Katz LA (2008). "The dynamic nature of eukaryotic genomes". Mol Biol Evol. 25: 787–794. doi:10.1093/molbev/msn032. PMC 2933061 . PMID 18258610. CS1 maint: Multiple names: authors list (link)
. PMID 18258610. CS1 maint: Multiple names: authors list (link)
^ Qiu Y.-L., Taylor A. B., McManus H. A. (2012). "Evolution of the life cycle in land plants". Journal of Systematics and Evolution. 50: 171–194. doi:10.1111/j.1759-6831.2012.00188.x. CS1 maint: Multiple names: authors list (link)
^ Crosland MW, Crozier RH (1986). "Myrmecia pilosula, an Ant with Only One Pair of Chromosomes". Science. 231 (4743): 1278. Bibcode:1986Sci...231.1278C. doi:10.1126/science.231.4743.1278. PMID 17839565.
^ "Archived copy" (PDF). Archived from the original (PDF) on 2014-02-23. Retrieved 2014-02-18.
^ Nuismer S.; Otto S.P. (2004). "Host-parasite interactions and the evolution of ploidy". Proc. Natl. Acad. Sci. USA. 101 (30): 11036–11039. Bibcode:2004PNAS..10111036N. doi:10.1073/pnas.0403151101. PMC 503737 .
.
^ Langlet, 1927.
^ Winge, 1917.
^ Manton, 1932.
^ Fabbri, F. 1963. Primo supplemento alle tavole cromosomiche delle Pteridophyta di Alberto Chiarugi. Caryologia 16: 237–335, [3].
^ http://mcb.berkeley.edu/courses/mcb142/lecture%20topics/Amacher/LECTURE_10_CHROM_F08.pdf
^ Gallardo MH, González CA, Cebrián I (2006). "Molecular cytogenetics and allotetraploidy in the red vizcacha rat, Tympanoctomys barrerae (Rodentia, Octodontidae)]". Genomics. 88 (2): 214–221. doi:10.1016/j.ygeno.2006.02.010. PMID 16580173.
^ Gallardo M. H.; et al. (2004). "Whole-genome duplications in South American desert rodents (Octodontidae)". Biological Journal of the Linnean Society. 82: 443–451. doi:10.1111/j.1095-8312.2004.00331.x.
^ Svartman, Marta; Stone, Gary; Stanyon, Roscoe (2005). "Molecular cytogenetics discards polyploidy in mammals". Genomics. 85 (4): 425–30. doi:10.1016/j.ygeno.2004.12.004. PMID 15780745.
^ http://web.uct.ac.za/depts/mmi/jmoodie/hiv2.html
^ Books, Elsevier Science & Technology (1950-01-01). Advances in Genetics. Academic Press. ISBN 978-0-12-017603-8.
^ Cosín, Darío J. Díaz, Marta Novo, and Rosa Fernández. "Reproduction of Earthworms: Sexual Selection and Parthenogenesis." In Biology of Earthworms, edited by Ayten Karaca, 24:69-86. Berlin, Heidelberg: Springer Berlin Heidelberg, 2011.
^ Dierschke T, Mandáková T, Lysak MA, Mummenhoff K (September 2009). "A bicontinental origin of polyploid Australian/New Zealand Lepidium species (Brassicaceae)? Evidence from genomic in situ hybridization". Annals of Botany. 104 (4): 681–688. doi:10.1093/aob/mcp161. PMC 2729636 . PMID 19589857.
. PMID 19589857.
^ Simon Renny-Byfield; et al. (2010). "Flow cytometry and GISH reveal mixed ploidy populations and Spartina nonaploids with genomes of S. alterniflora and S. maritima origin". Annals of Botany. 105 (4): 527–533. doi:10.1093/aob/mcq008. PMC 2850792 . PMID 20150197.
. PMID 20150197.
^ ab Kim E. Hummer; et al. (March 2009). "Decaploidy in Fragaria iturupensis (Rosaceae)". Am. J. Bot. pp. 713–716. doi:10.3732/ajb.0800285.
^ Talyshinskiĭ, G. M. (1990). "Study of the fractional composition of the proteins in the compound fruit of polyploid mulberry". Shelk (5): 8–10.
^ Fujikawa-Yamamoto K (2001). "Temperature dependence in Proliferation of tetraploid Meth-A cells in comparison with the parent diploid cells". Cell Structure and Function. pp. 263–269. doi:10.1247/csf.26.263.
^ Kiichi Fukui; Shigeki Nakayama. Plant Chromosomes: Laboratory Methods.
^ "Genes involved in tissue and organ development: Polytene chromosomes, endoreduplication and puffing". The Interactive Fly.
^ Ramsey, J.; Schemske, D. W. (2002). "Neopolyploidy in Flowering Plants" (PDF). Annual Review of Ecology and Systematics. 33: 589–639. doi:10.1146/annurev.ecolsys.33.010802.150437.
^ Hansen MT (1978). "Multiplicity of genome equivalents in the radiation-resistant bacterium Micrococcus radiodurans". J. Bacteriol. 134 (1): 71–5. PMC 222219 . PMID 649572.
. PMID 649572.
^ Soppa J (2011). "Ploidy and gene conversion in Archaea". Biochem. Soc. Trans. 39 (1): 150–4. doi:10.1042/BST0390150. PMID 21265763.
^ Zahradka K, Slade D, Bailone A, Sommer S, Averbeck D, Petranovic M, Lindner AB, Radman M (2006). "Reassembly of shattered chromosomes in Deinococcus radiodurans". Nature. 443 (7111): 569–73. Bibcode:2006Natur.443..569Z. doi:10.1038/nature05160. PMID 17006450.
^ Kottemann M, Kish A, Iloanusi C, Bjork S, DiRuggiero J (2005). "Physiological responses of the halophilic archaeon Halobacterium sp. strain NRC1 to desiccation and gamma irradiation". Extremophiles. 9 (3): 219–27. doi:10.1007/s00792-005-0437-4. PMID 15844015.
^ Schaechter, M. Eukaryotic microbes. Amsterdam, Academic Press, 2012, p. 217.
^ Edwards MJ; et al. "Mixoploidy in humans: two surviving cases of diploid-tetraploid mixoploidy and comparison with diploid-triploid mixoploidy". Am J Med Genet. 52: 324–30. doi:10.1002/ajmg.1320520314. PMID 7810564.
^ Järvelä, IE; Salo, MK; Santavuori, P; Salonen, RK. "46,XX/69,XXX diploid-triploid mixoploidy with hypothyroidism and precocious puberty". J Med Genet. 30: 966–7. doi:10.1136/jmg.30.11.966. PMC 1016611 . PMID 8301657.
. PMID 8301657.
^ Bender K (1963). "Über die Erzeugung und Entstehung dihaploider Pflanzen bei Solanum tuberosum"". Zeitschrift für Pflanzenzüchtung. 50: 141–166.
^ Nogler, G.A. 1984. Gametophytic apomixis. In Embryology of angiosperms. Edited by B.M. Johri. Springer, Berlin, Germany. pp. 475–518.
^ * Pehu E (1996). "The current status of knowledge on the cellular biology of potato". Potato Research. 39: 429–435. doi:10.1007/bf02357948.
^ * Sprague G.F.; Russell W.A.; Penny L.H. (1960). "Mutations affecting quantitative traits in the selfed progeny of double monoploid maize stocks". Genetics. 45 (7): 855–866.
^ Pandit, M. K.; Pocock, M. J. O.; Kunin, W. E. (2011-03-28). "Ploidy influences rarity and invasiveness in plants". Journal of Ecology. Wiley-Blackwell. 99: 1108–1115. doi:10.1111/j.1365-2745.2011.01838.x.
^ Gilbert, Natasha (2011-04-06). "Ecologists find genomic clues to invasive and endangered plants". nature.com. Nature Publishing Group. doi:10.1038/news.2011.213. Retrieved 2011-04-07.
Sources
- Griffiths, A. J. et al. 2000. An introduction to genetic analysis, 7th ed. W. H. Freeman, New York ISBN 0-7167-3520-2
External links
Some eukaryotic genome-scale or genome size databases and other sources which may contain the ploidy of many organisms:
- Animal genome size database
- Plant genome size database
- Fungal genome size database
Protist genome-scale database of Ensembl Genomes
Nuismer S.; Otto S.P. (2004). "Host-parasite interactions and the evolution of ploidy". Proc. Natl. Acad. Sci. USA. 101: 11036–11039. Bibcode:2004PNAS..10111036N. doi:10.1073/pnas.0403151101. PMC 503737 . (Supporting Data Set, with information on ploidy level and number of chromosomes of several protists)
. (Supporting Data Set, with information on ploidy level and number of chromosomes of several protists)
Chromosome number and ploidy mutations YouTube tutorial video